I can recommend Kai Kupferschmidt’s article here at Science for background. The short form is that earlier this month, UK authorities noticed that there was an upswing in reported SARS-CoV-2 cases in southeast England, and that sequencing was showing that these seemed to be tied to a new variant of the virus itself. (As Kai’s article notes, there’s a feature of the PCR testing that made it easier to pick this out). There have been cases in several other countries as well.
If you’re counting back to the original Wuhan type, this one has piled up 17 mutations, which is an unusually high number, for sure. Let’s take a look at those in the context of the whole virus’ sequence – below is the “Events” view from Nextstrain.org, which shows you how many reports they’ve had of mutations at particular amino acids.
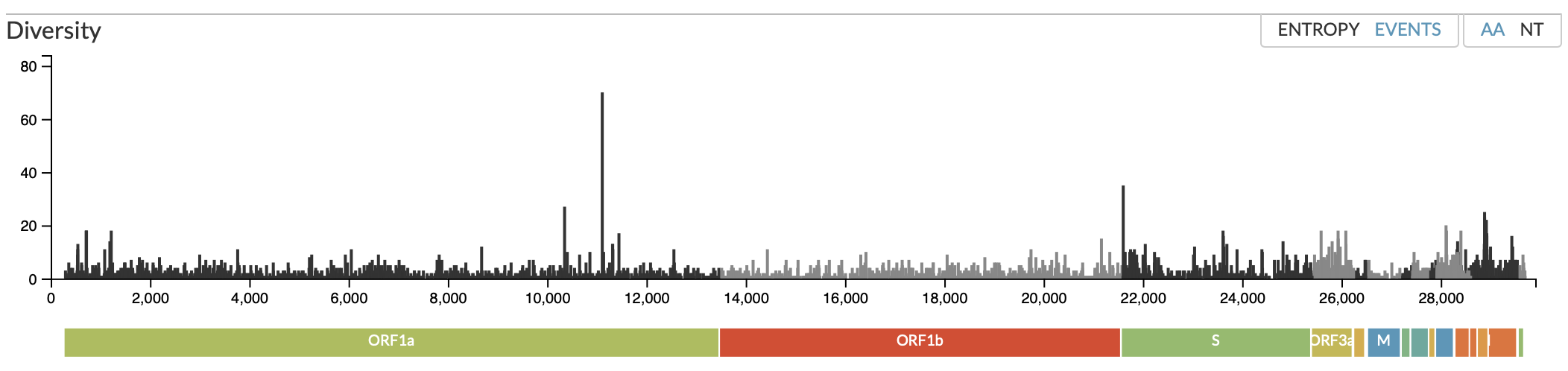
Virus aficionados will know immediately what they’re looking at here, but if that diagram looks odd to you, here’s how to read it. The vertical-black-line part represents number of reported mutations in that particular three-letter “codon”, which is the unit of DNA/RNA that codes for one amino acid in the resulting protein. That tallest line, for example, represents 70 mutations that have turned up in a particular codon that codes for an amino acid in the viral ORF1a protein. You can see the associated protein by the color of the bar underneath it and its label. That bar is the whole coronavirus genome, laid out in terms of its various proteins. The (in)famous Spike protein is the third biggest, third in line: you go through the ORF1a protein, then ORF1b, and then you have S for Spike (the second green region). And you can see that there are plenty of reported mutations there – more than (say) in the ORF1b protein right before it , which is relatively quiet. So it’s not like we haven’t been seeing Spike mutations; they’re happening all the time.
Now, these lines in the chart are single changes, but as the virus rolls along, it piles mutations on top of mutations. The order that these occur in lets you put together a “tree” diagram based on the branching order in which these took place, and Nextstrain is a great place to see those in all their glory. When you look at the 17-mutation strain that’s of concern in England, several things stand out, as analyzed in this preprint. For one thing, all of these mutations are ones that lead to a different amino acid when that particular codon is read off. The three-letter code has some redundancies in it; some of the changes you can make will lead to the same amino acid in the end, but quite a few of the changes in this latest strain are real change-the-protein ones – there are 14 of those and three that lead to deletions, along with six silent ones that don’t change anything in the end. This strain is in Clade 20B on the Nextstrain charts, and there are charts here that will show you the relationships in detail.
There are three of these amino acid changes in the ORF1a and ORF1b regions, and one deletion, two deletions and six amino acid changes in the Spike protein, three more mutations in Orf8, and two changes in the nucleotide (N) protein. Three of the Spike region mutations have been already described as having significance for its function: the N501Y is on the of the key receptor-binding-domain amino acids at the very tip of the Spike, the part that contacts the human ACE2 protein. (Here are some new data that should make us a bit happier, though: this mutation does not seem to be associated with loss of neutralization when exposed to human antibodies) One of the deletions (69-70del) has shown up several times in other SARS-CoV-2 sequences (including a mink-related outbreak in Denmark), and is speculated to be involved in evading immune response (to some degree). And the P681H is right next to the “furin cleavage” site, which is known to be key for the process of virus going on to enter human cells.
According to that paper last linked above, this list suggests some real selection pressure is at work here, not just random mutational drift and wandering. As mentioned, several of these have shown up independently in the past, but this combination of them is new. The authors speculate that these multiple changes might have occurred, at least some of them, in an individual with long-term infection, where the virus had a chance to deal with the human immune system for an extended period. It’s a plausible idea, because something different had to happen to allow so many mutations to pile up more or less at once, but it remains unproven. It should be noted that not all of the mutations seen in this strain are known to be trouble: there’s a stop codon mutation in ORF8, and something like it was seen earlier in Singapore in a branch of SARS-CoV-2 that later died out thanks to that country’s stringent control measures. It seemed to be associated with milder infection, though its significance when combined with these other mutations in unknown. But as the Kupferschmidt article details, there are still a lot of explanations in play. The increase of this strain in the UK could certainly be down to higher infectiousness, but we’ve thought that about other strains in the past, and it hasn’t panned out. It’s in the “remains to be seen” category, because the vagaries of person-to-person transmission (and the vagaries of human movement!) can produce apparent patterns that aren’t what they appear to be.
I should note here that there’s another strain in South Africa that is bringing on similar concerns. This one has eight mutations in the Spike protein, with three of them (K417N, E484K and N501Y) that may have some functional role. You’ll note the N501Y is also in the UK strain, but the E484K is one that people are particularly watching, because that’s in a region that a number of antibodies seem to recognize. The South African one also seems to be moving up when you look at population sequencing data, suggesting that it might also be more transmissible.
But remember, antibody response (whether through infection or through getting vaccinated) is polyclonal: you raise a number of different antibodies that bind in different ways. That’s one of the key features of the adaptive immune system: hitting a new antigen from a number of directions at once. This makes it harder to slip through for a new pathogen variant – which is good, but at the same time, it’s not impossible, either. Thus the attention being paid to these two new strains.
That leads us to the other big factor that people are worried about: increased transmissibility is not good, of course, but what happens after you’ve been infected? Do these strains produce the same sort of disease in humans? This is completely unclear at this point – so far, no one is associating either of these variants with a notably worse clinical course of disease, though. Many infectious pathogens, in fact, gradually evolve versus a given animal host to be more infectious and less virulent over time. Remember, it’s not the job of a virus to make people deathly ill: it’s the job of a virus to make more virus. Overall, that is generally better served by strains that are easier to catch and that don’t rip into their hosts too viciously. But this is a big-picture statistical effect, and not necessarily at work in any given strain that emerges. There could be a new strain that is both more infectious and more harmful once caught, and we have to keep our guard up against such a thing. Even a more transmissible virus that produces the same level of illness would be (of course) a very bad thing.
So I think the amount of scientific attention being paid to these new strains is completely appropriate. The popular press might be another story. It’s important to remember that (as mentioned) we haven’t even established for sure that these strains are in fact more infectious, and that we don’t know a thing yet for sure about what effect they have in humans compared to the other variants. So if you see any headlines about Relentless March of the Supervirus, go read something else, because that stuff is (fortunately) way out ahead of the facts on the ground. These are by no means the last variants like these that we’re going to be seeing, and we need to learn how to cover them in a responsible way.
So what’s coming next, and when we will know more? We will have animal-model data coming soon to tell us something about infectiousness, and there are already studies underway using human antibody mixtures (from infected patients and vaccinated ones) to see if these new strains are any less susceptible to our immune response. This will be a matter of weeks; I wouldn’t expect to see any clarity before then. And that’s also at least the time scale we would need to start confirming the clinical effects in the human population – you can be sure that medical centers around the world will be monitoring patients who have been confirmed with these variants to see if there are any differences. We’ll also want to know how these look in different age cohorts, in people with pre-existing conditions, and so on, but all of this will take irreducible amounts of time to get a meaningful picture.
My speculations are worth what you’re paying for them. But I think that odds are reasonable that UK strain, based on what we’re seeing so far, may well be more infectious than the existing ones. I hope I’m wrong about that, and I want to re-emphasize that I very well could be. At the same murky level of clarity, I’m not seeing anything so far that makes me think that it causes a worse form of the disease, and I very much hope that I’m not wrong about that. As for vaccine effects, my money is on the antibody response from the vaccines still being protective – and that’s going to be some of the first hard data that we get, because those are some of the most straightforward experiments to run. Updates as we get them.
